Presenter Name: Aaron Bohn
Description
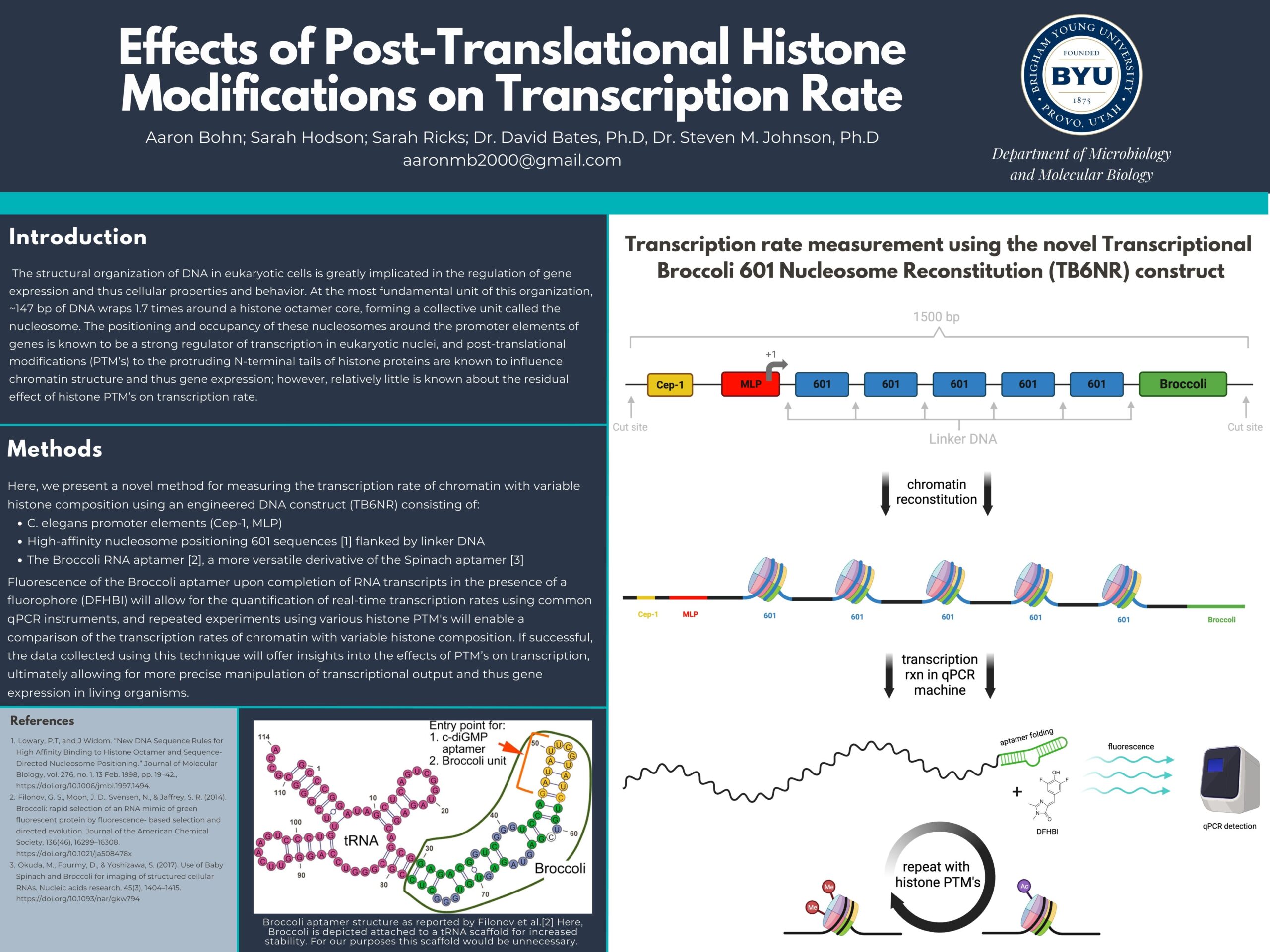
The structural organization of DNA in eukaryotic cells is highly implicated in the regulation of gene expression and thus cellular properties and behavior. At the most fundamental unit of this organization, approximately 147 base pairs of DNA wraps about 1.7 times around a histone octamer core, forming a collective unit called the nucleosome. The positioning and occupancy of these nucleosomes around the promoter elements of genes is known to be a strong regulator of transcription in eukaryotic nuclei, and post-translational modifications (PTM's) to the protruding N-terminal tails of histone proteins are known to influence chromatin structure and thus gene expression; however, relatively little is known about the residual effect of histone PTM's on transcription rate. Here, we present a novel method for measuring the transcription rate of chromatin with variable histone composition using an engineered DNA construct consisting of C. elegans promoter elements and high-affinity nucleosome positioning sequences1 followed by the Broccoli2 aptamer, a more versatile derivative of the Spinach3 aptamer. This terminal RNA aptamer will bind a DFHBI fluorophore molecule and fluoresce upon completion of RNA transcripts, allowing for the quantification of real-time transcription rates using common qPCR instruments. After using salt dialysis to reconstitute chromatin in vitro from the engineered DNA construct and histones with varying PTM's, this technique will enable us to measure the hypothesized changes in transcription rate as histones in the gene body are differentially modified. If successful, the data collected using this technique will offer insights into the effects of PTM's on transcription rate, ultimately allowing for more precise manipulation of transcriptional output and thus gene expression in living organisms.
Literature Cited:
1. Lowary, P.T, and J Widom. "New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-Directed Nucleosome Positioning." Journal of Molecular Biology, vol. 276, no. 1, 13 Feb. 1998, pp. 19-42., https://doi.org/10.1006/jmbi.1997.1494.
2. Filonov, Grigory S., et al. "Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution." Journal of the American Chemical Society, vol. 136, no. 46, 22 Oct. 2014, pp. 16299-16308., https://doi.org/10.1021/ja508478x.
3. Paige, Jeremy S., et al. "RNA Mimics of Green Fluorescent Protein." Science, vol. 333, no. 6042, 2011, pp. 642-646., https://doi.org/10.1126/science.1207339.
1. Lowary, P.T, and J Widom. "New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-Directed Nucleosome Positioning." Journal of Molecular Biology, vol. 276, no. 1, 13 Feb. 1998, pp. 19-42., https://doi.org/10.1006/jmbi.1997.1494.
2. Filonov, Grigory S., et al. "Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution." Journal of the American Chemical Society, vol. 136, no. 46, 22 Oct. 2014, pp. 16299-16308., https://doi.org/10.1021/ja508478x.
3. Paige, Jeremy S., et al. "RNA Mimics of Green Fluorescent Protein." Science, vol. 333, no. 6042, 2011, pp. 642-646., https://doi.org/10.1126/science.1207339.
University / Institution: Brigham Young University
Type: Poster
Format: In Person
Presentation #B63
SESSION B (10:45AM-12:15PM)
Area of Research: Science & Technology
Email: aaronmb2000@gmail.com
Faculty Mentor: Steven Johnson